Motivation: A rapid discovering of experimental data on regulation of gene expression and their accumulation in various databases accompanied by development of numerous and diverse software tools for their analyses demands integration of the available informational and software resources.
Introduction
Currently, the number of databases on gene expression and a variety of software for the analysis of these data are growing fast. An Internet-accessible system GeneExpress-2.1 is being developed for accumulation of experimental data, their analysis, and navigation through integrated software and informational resources related to regulation of gene expression. It integrates a large amount of databases and hundreds of programs for processing the data on the structure-function organization of DNA, RNA, proteins, and gene networks together with other informational resources important for Informational resources of GeneExpress.
The structure
of the GeneExpress-2.1 corresponds
to the natural hierarchical organization of molecular genetic systems,
containing the following levels: (1) DNA
level, (2) RNA level, (3) protein level, and (4) gene network level. Each module
contains (1) experimental data represented as a database or a sample; (2) program
for data analysis; (3) results of an automated data processing; and (4) tools
for graphical representation of these data and results of the data analyses.
The main databases of GeneExpress-2.1 involve the relational model of data
representation. For access to the
databases of GeneExpress-2.1 system, RDBMS (Relation Database Management
System), ORACLE 9i, and Sequence Retrieval System (SRS 6.0) are used.
Research into mechanisms underlying molecular interactions depending on the genetic information and specific features of molecular structures may shed the light on biochemical functions and roles of elementary components as well as on the specific control patterns of gene networks. These pieces of knowledge form the background for computer simulation of gene networks allowing changes in molecular genetic, biochemical, physiological, morphological, and other characteristics of various organisms to be predicted as well as optimal control actions and stimuli for correcting genetically specified impairments of the body operation to be searched for.
For this purpose, a new generation computer technologies integrated in the computer system GeneNetDiscovery is developed at the Siberian Branch of the Russian Academy of Sciences. This system provides solving a wide range of problems in the field of computer analysis and simulation of complex molecular genetic systems (gene networks, genetically controlled metabolic pathways, signal transduction pathways, etc.) including (i) accumulation of data and knowledge on the structure-function organization of gene networks; (ii) integration of the information on gene networks and metabolic pathways; (iii) construction of gene network mathematical models and their computer-assisted numerical analysis; (iv) study of dynamic behavior of complex molecular genetic systems (gene networks) in norm, in case of pathologies and metabolic diseases, and under the effect of adverse environmental factors at molecular genetic, cellular, and organismal levels; and (v) search for optimal control of gene networks and correction of their behavior in the case of various pathological states.
1.
Transcription
Regulatory Regions Database (TRRD)
TRRD is designed for accumulation of experimental information on the structure-function organization of regulatory regions of eukaryotic genes. It is a unique informational resource on long gene transcription regulatory regions. In addition to description of regulatory region itself, it provides (a) description of the hierarchy of all the regulatory units included into a described regulatory region (such as transcription factor binding sites, promoters, enhancers, silencers, etc.); (b) information on expression patterns of the genes described; and (c) information on physiological systems, organs, and cell types wherein these genes are expressed. The new release of TRRD-6.0 contains interferon-inducible genes; erythroid-specific genes; genes of lipid metabolism in liver, adipose tissue, at the cell and organismal levels (cholesterol regulation, and leptin hormone regulation, lipid exchange between lipoprotein blood particles); glucocorticoid-inducible gene;, cell cycle-dependent genes; genes of the endocrine system; heat shock-regulated genes; redox sensitive genes; iron metabolism genes; macrophage-expressed genes; apoptosis genes; and plant genes. TRRD-6.0 comprises descriptions of 1405 genes, 6646 sites, and 2158 regulatory regions. This database is supported constructed using ORACLE 9i, and Sequence Retrieval System (SRS 6.0) is used to access TRRD-6.0. A novel version of the TRRD Viewer (release 2.0) implemented as a Java-applet allows the regulatory gene regions described in TRRD to be visualized.gene expression regulation. From its first version (Kolchanov at la., 1998a; 1998b), GeneExpress is intensively developing. This paper briefs the state of GeneExpress –2.1 in 2002. Descriptions of its individual modules in more detail are available in papers included in Proceedings of BGRS’2002. The system is widely and actively used for computer analyses of various organizational levels of molecular genetic systems.
2. Programs for recognizing regulatory elements involved in controlling the transcription
GeneExpress-2.1 has a large set of original programs for recognition and various analyses of transcription factor binding sites and promoters as well as study of specific contextual and structural DNA features in gene regulatory regions, exemplified below.
Resource |
Description |
|
BinomSite |
Searching for potential transcription factor
binding sites (TFBS) using a binomial criterion for estimating similarity
scores between regions of a sequence analyzed and the TFBS sequences
described in TRRD URL: http://wwwmgs.bionet.nsc.ru/mgs/programs/mmsite/ |
|
|
Simultaneous usage of the entire set of
recognition methods for detecting TFBSs. URL: http://wwwmgs.bionet.nsc.ru/mgs/programs/multalig/ |
|
ARGO_Viewer |
Recognition of promoters of
tissue-specific gene groups basing on the analysis of the presence of
specific quasi-invariant oligonucleotide motifs detected using the program ARGO. URL: http://wwwmgs.bionet.nsc.ru/mgs/programs/argo/argo_viewer.html
|
|
RGSiteScan |
Searching for TFBSs basing on the recognition group
approach. URL: http://wwwmgs.bionet.nsc.ru/mgs/programs/yura/RecGropScanStart.html
|
|
KD_Prom |
Recognition of RNA PolII
promoters from their contextual patterns determined using knowledge discovery
and data mining in TRRD. URL: http://wwwmgs.bionet.nsc.ru/mgs/programs/recon2/
|
|
ProGA |
Recognition of RNA PolII
promoters. URL: http://wwwmgs.bionet.nsc.ru/mgs/programs/proga/
|
|
BLAST_Promoter |
Recognition of the RNA PolII
promoters basing on the BLAST search for homology with the promoters described
in the TRRD. URL: http://wwwmgs.bionet.nsc.ru/mgs/systems/fastprot/blast.html
|
|
Recon |
Searching
for potential nucleosome formation sites basing on the nonuniformity of
dinucleotide context within local promoter regions. URL: http://wwwmgs.bionet.nsc.ru/mgs/programs/recon/
|
3.
Informational
and software resources on RNA structure-function organization
To solve a variety of problems on analyzing RNA structure-function organization, a number of databases and software tools were developed with GeneExpress-2.1 and united in the module RNA Integration Level. It comprises (i) a number of programs for calculation of RNA secondary structure and evaluation of the secondary structure formation potential and (ii) the knowledge base on structure-function organization of leader mRNA sequences.
Resource |
Description |
|
Program GArna |
Applying genetic
algorithm to predict the secondary structures displaying seals energies and
visualize them. URL:
http://wwwmgs.bionet.nsc.ru/mgs/programs/2dstructrna/
|
|
Program MatrixSS |
Calculation of E score, a contextual characteristics reflecting the potential for forming RNA secondary structure compared with random sequences. URL: http://wwwmgs.bionet.nsc.ru/mgs/programs/2dstructrna/MatrixSS.html |
|
Knowledge base LEADER_RNA |
LEADER_RNA is a tool
to evaluate mRNA translational properties. Contains a database with samples
of 5'UTR sequences of high- and low-expressed mRNAs of mammals, dicot, and
monocot plants. These sequences are used as training samples for the computer
system. This knowledge base contains also (1) description of the discovered
mRNA properties that may be used to discriminate between the high- and
low-expressed mRNAs and (2) programs predicting mRNA translational
efficiencies from significant contextual and structural characteristics of
mRNA 5'UTRs (C codes for prediction of mRNA translation level). URL:
http://wwwmgs.bionet.nsc.ru/mgs/gnw/leader/
|
4.
Informational and software
resources on the structure-function organization of proteins
A number of databases and software tools, forming the modules Protein Integration Level of GeneExpress-2.1, have been developed for solving the problems related to analyses, structure, function, and evolution of proteins. This module contains the databases on (i) expanded annotation of the EnPDB-compiled structures, (ii) active sites of the PDBSite-compiled proteins, (iii) protein and peptide sequences obtained by artificial in vitro selection (ASPD) as well as the programs for (iv) searching the protein spatial structure for re regions similar to PDBSiteScan-compiled active centers and (ii) detecting and analyzing the coordinatively fixed amino acid substitutions (CRASP, Gene Network Level)
Resource |
Description |
|
Database EnPDB |
Expanded options for indexed search for
information in the PDB databank entries URL: http://wwwmgs.bionet.nsc.ru/mgs/gnw/enpdb/ |
|
Database PDBSite |
Information on the
spatial structure and physicochemical properties of 4723 active protein sites
annotated in PDB. URL: http://wwwmgs.bionet.nsc.ru/mgs/gnw/pdbsite/ |
|
Program PDBSiteScan |
Searching for active
sites in protein spatial structures according to the sequence and spatial
arrangement of amino acid residues. URL: http://wwwmgs.bionet.nsc.ru/mgs/systems/fastprot/pdbsitescan.html |
|
Database ASPD |
Information on peptide
and protein sequences produced an in vitro selection. URL: http://wwwmgs.bionet.nsc.ru/mgs/gnw/aspd/ |
|
Program CRASP |
Detection of
coordinated amino acid substitutions in protein families and analysis of
their physicochemical characteristics. URL: http://wwwmgs.bionet.nsc.ru/mgs/gnw/crasp/ |
5.
Informational and software resources on the structure-function organization and
operation dynamics of gene
networks
This module of GeneExpress-2.1, GeneNetDiscovery, is a tool designed for
creating gene network models that could be further used in another operation
system and for a variety of purposes.
For annotators and modelers, specialized worksites are developed in a
Windows NT/2000/XP environment. Web interfaces are developed for outside
“casual” users. Currently, two client programs are realized (gene network
editor and gene network viewer), which communicate with the server by HTTP
protocol realized as exchange by the XML messages.over HTTP protocol, by which XML
messages are sent. Oracle9i is appled for controlling the databases. When
developing the system’s middleware controlling the logics of its operation and
its linkage to the knowledge bases and databases, application server Oracle9iAS
is used; it includes Container
for J2EE (OC4J), XML SQL Utility, and XML Parser as components. The
operation logics of the system GeneNetDiscovery is shown in Fig. 1.
Fig.
1. Functional layout of the system GeneNetDiscovery. The system
GeneNetDiscovery comprises the following functional modules: (1) Subsystem
for constructing models, including A new version
of this database in an Oracle9i
environment is under development now (Loktev et al., 2002); (2)
Subsystem for analyzing gene network models, including (3)
Subsystem for identifying models, including (4)
Subsystem for simulating gene networks and analyzing their
behavior, including URL: http://wwwmgs.bionet.nsc.ru/mgs/gnw/gn_model/modelling.shtml Sets of differential
equations describing dynamics of reactions, reaction rate constants, and
initial concentrations of the components for dynamical models of three gene
networks, namely, gene
network regulating (1) lipid
metabolism, (2) erythrocyte differentiation and maturation under the effect of
EPO, and (3) activation of macrophages by IFN-g
and LPS. URL: http://wwwmgs.bionet.nsc.ru/mgs/gnw/gn_model/ (5)
Subsystem for controlling gene networks, including Described in
the work is the architecture of the system that is developed basing on
multilevel computational approaches combining genome-encoded information with
nongenomic network connections. A part of the modules of the system
GeneNetDiscovery has been realized. The major algorithms forming the core of
this system have been tested by solving particular problems. Acknowledgements Work was supported in part by the Russian
Foundation for Basic Research (grants No. 01-07-90376, 01-07-90084, 00-07-90337,
02-07-90355, 02-07-90359, 00-04-49229, and 00-04-49255); Russian Ministry of
Industry, Science, and Technologies (grant No. 43.073.1.1.1501); Siberian
Branch of the Russian Academy of Sciences (Integration Projects Nos. 65 and 66)
US National Institutes of Health (grant No. 2 R01-HG-01539-04A2); and US
Department of Energy (grant No. 535228 CFDA 81.049). References 1. Kolchanov,
N.A., Ponomarenko, M.P., Kel, A.E., Kondrakhin, Yu.V., Frolov, A.S., Kolpakov,
F.A., Kel, O.V., Ananko, E.A., Ignatieva, E.V., Podkolodnaya, O.A., Stepanenko,
I.L., Merkulova, T.I., Babenko, V.N., Vorobiev, D.G, Lavryushev, S.V.,
Ponomarenko, Yu.V., Kochetov, A.V., Kolesov, G.B., Podkolodny, N.L., Milanesi,
L., Wingender, E., Heinemeyer, T., and Solovyev, V.V. (1998a). GeneExpress: a computer system for description,
analysis, and recognition of regulatory sequences of the eukaryotic genome.
ISMB,. 6:95-104. MEDLINE PMID: 9783214; UI: 98456543. 2. Kolchanov,
N.A., Ponomarenko, M.P., Kondrakhin, Yu.V., Frolov, A.S., Kolpakov, F.A., Kel,
A.E., Kel-Margoulis, O.V., Ananko, E.A., Ignatieva, E.V., Podkolodnaya, O.A.,
Stepanenko, I.L., Merkulova, T.I., Babenko, V.N., Vorobiev, D.G., Lavryushev,
S.V., Grigorovich, D.A., Ponomarenko, J.V., Kochetov, A.V., Kolesov, G.B.,
Podkolodny, N.L., Wingender, E., Heinemeyer, T., Milanesi, L., Solovyev, V.V.,
and Overton, O.K. (1998b). GeneExpress system: description, analysis,
and recognition of regulatory sequences in eukaryotic genomes. Proc. I
Intern. Conference on Bioinformatics of Genome Regulation and Structure,
BGRS’98, Novosibirsk–Altai Mountains, Russia, August 24-31, 1998, 71-76. 3. Afonnikov, D.A. (2002). Contribution of
coadaptive substitutions to the stability of physicochemical properties of
ATP-binding sites in protein kinases. Proc. III Intern. Conference on
Bioinformatics of Genome Regulation and Structure (BGRS'2002). 4. Afonnikov,
D.A., Oshchepkov, D.Yu., and Kolchanov, N.A. (2001). Detection of conserved
physico-chemical characteristics of proteins by analyzing clusters of positions
with coordinated substitutions. Bioinformatics, 17, 1035-46. 5. Ananko,
E.A., Podkolodny, N.L., Ignatieva, E.V., Podkolodnaya, O.A., Stepanenko, I.L.,
and Kolchanov, N.A. (2002). GeneNet system: its status in 2002. Proc. III
Intern. Conference on Bioinformatics of Genome Regulation and Structure
(BGRS'2002). 6. Dobrynin,
A.A., Makarov, L.I., and Podkolodny, N.L (2002). A graph-theoretic approach to
computer analysis of gene network structure. Proc. III Intern. Conference
on Bioinformatics of Genome Regulation and Structure (BGRS'2002). 7. Loktev,
K.A., Tkachev, Yu.A., Ananko, E.A., and Podkolodny N.L. (2002). A system for
visual modeling of gene networks structural and functional. Proc. III Intern.
Conference on Bioinformatics of Genome Regulation and Structure (BGRS'2002). 8. Likhoshvai,
V.A., Latypov, A.F., Nedosekina, E. A., Ratushny, A.V., and Podkolodny, N.L.
(2002). Technology of using experimental data for verification of models of
gene network operation dynamics. Proc. III Intern. Conference on Bioinformatics
of Genome Regulation and Structure (BGRS'2002). 9. Borisova,
I.A., Zagoruiko, N.G., Likhoshvai, V.A., Ratushny, A.V., and Kolchanov, N.A.
(2002). Diagnostics of mutations based on analysis of gene networks. Proc. III
Intern. Conference on Bioinformatics of Genome Regulation and Structure
(BGRS'2002). 10. Fadeev, S.I.,
Berezin, A.Yu., Gainova, I.A., Kogai, V.V., Ratushny, A.V., and Likhoshvai,
V.A. (2002). Development of the program software for mathematic modeling of the
gene network dynamics . Proc. III Intern. Conference on Bioinformatics of
Genome Regulation and Structure (BGRS'2002). 11. Kudryavtseva, A.N.
and Stepanenko, I.L. (2002). Gene network of glutathione homeostasis: a
response to oxidation stress. Proc. III Intern. Conference on Bioinformatics of
Genome Regulation and Structure (BGRS'2002). 12. Latypov, A.F.,
Nikulichev, Yu.V., Likhoshvai, V.A., Ratushny, A.V., Matushkin, Yu.G., and
Kolchanov N.A. (2002a). A method of solving problems of optimal control in
dynamics of gene networks. Proc. III Intern. Conference on Bioinformatics of
Genome Regulation and Structure (BGRS'2002). 13. Latypov, A.F.,
Nikulichev, Yu.V., Likhoshvai, V.A., Ratushny, A.V., Matushkin, Yu.G., and
Kolchanov N.A. (2002b). Problems of control of gene networks in a space of
stable states. Proc. III Intern. Conference on Bioinformatics of Genome
Regulation and Structure (BGRS'2002). 14. Nedosekina, E.A.
and Ananko, E.A. (2002). Gene network of macrophage activation under the action
of interferon-gamma and lipopolysaccarides. Proc. III Intern. Conference on
Bioinformatics of Genome Regulation and Structure (BGRS'2002). 15. Ratushny, A.V. and
Likhoshvai, V.A. (2002). Computer analysis of the effects of mutations in LDL
receptor gene on the regulation of cholesterol biosynthesis in the cell. Proc.
III Intern. Conference on Bioinformatics of Genome Regulation and Structure
(BGRS'2002). 16. Ratushny, A.V.,
Likhoshvai, V.A., and Kolchanov, N.A. (2002). Analysis of mutational portraits
of gene networks. Proc. III Intern. Conference on Bioinformatics of Genome
Regulation and Structure (BGRS'2002). 17. Turnaev, I.I. and
Podkolodnaya, O.A. (2002). Gene network on cell cycle control. Proc. III
Intern. Conference on Bioinformatics of Genome Regulation and Structure
(BGRS'2002). 18. Stepanenko, I.L.
and Grigor’ev, S.A. (2002). Organization of the gene network of apoptosis.
Proc. III Intern. Conference on Bioinformatics of Genome Regulation and
Structure (BGRS'2002).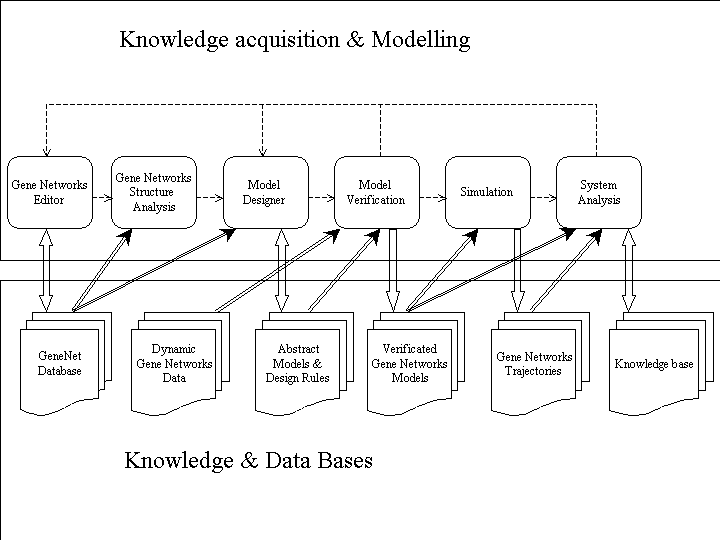
Conclusion



![[SBRAS]](/gif/s_sigma.gif)